On the bottom toolbar of the main form, click the Examples button. With the Examples folder open, navigate to the DrPhylo sub-folder and select the Rooted_Fungi_Tree.nwk file (if this file is not visible, set the filter to All Files)
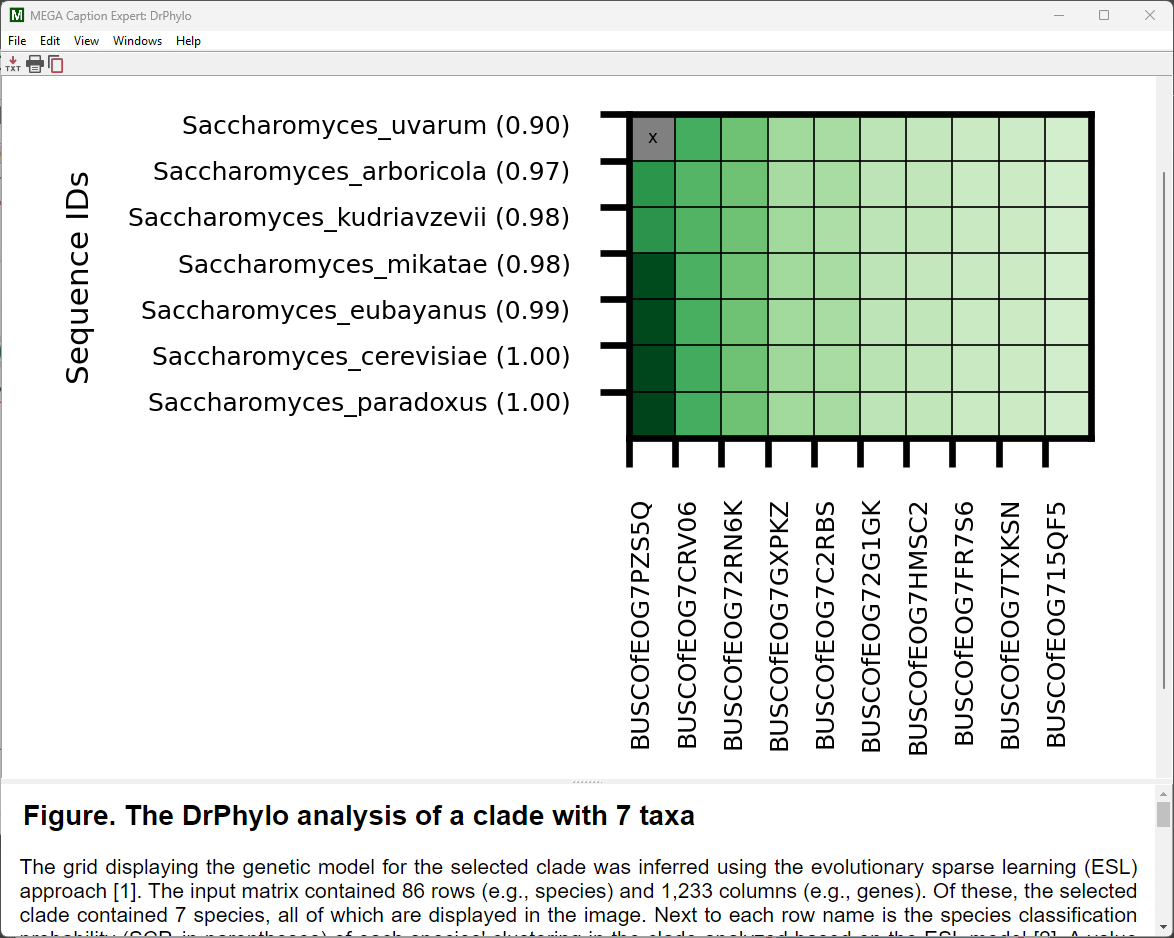
The tree of Fungi will be displayed in the Tree Explorer window. Right-click the branch for the Saccharomyces clade and select Launch DrPhylo from the context-menu.
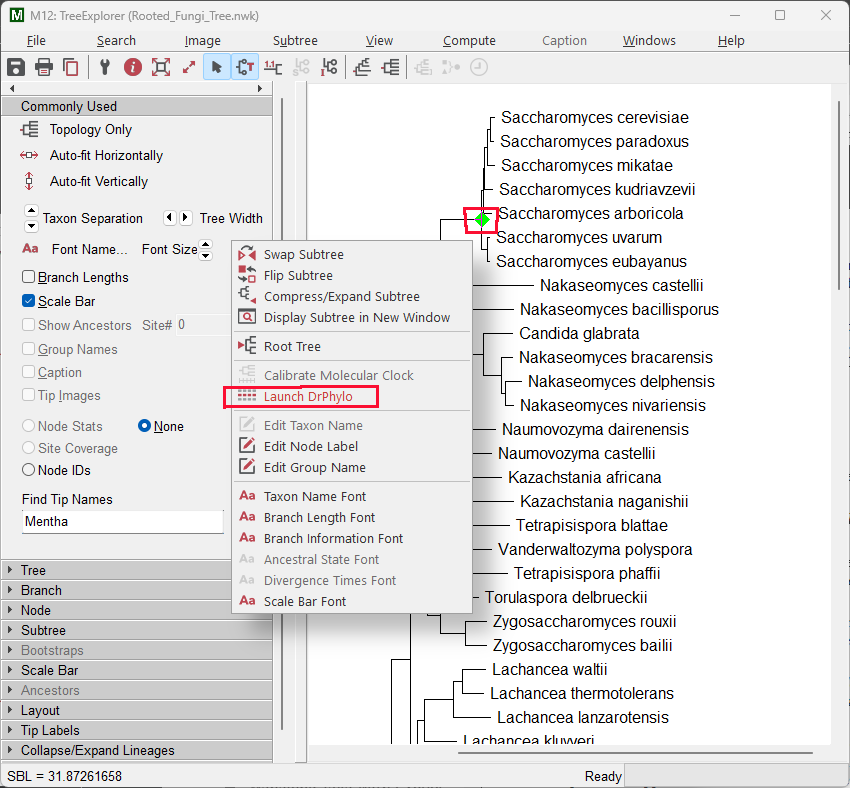
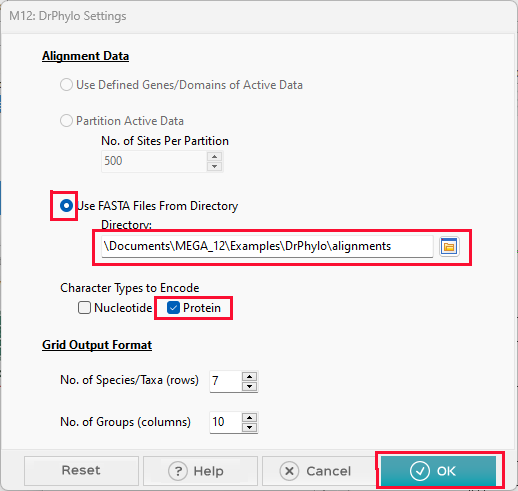
In the DrPhylo Settings window that is displayed, click the Use FASTA Files From Directory radio button and then click the folder icon. In the Select Folder window that is displayed, select the ~/Documents/Examples/DrPhylo/alignments folder that is included with the MEGA installation. This directory contains aligned FASTA files for each gene of interest.
Under Character Types to Encode, click the Protein check box.
Click the OK button to launch the analysis.