In real data, amino acid frequencies usually vary among different kinds of amino acids. In this case, a correction based on the equal input model gives a better estimate of the number of amino acid substitutions than does the Poisson correction distance. Note that this assumes an equality of substitution rates among sites. When the amino acid frequencies are different between the sequences, the modified formula (Tamura and Kumar 2002) relaxes the estimation bias.
MEGA provides facilities for computing the following quantities:
|
Quantity |
Description |
|
d: distance |
Number of amino acid substitutions per site. |
|
L: No of valid common sites |
Number of sites compared. |
Formulas used are:
Distance
![]()
where p is the proportion of different amino acid sites, gXi is the frequency of amino acid i for sequence X, gi is the average frequency for the pair of the sequences, and
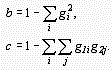
The variance of d can be estimated by the bootstrap method.